In this two part series, I want to examine how genetically identical cells in equ
 al environments can undergo very different developmental changes. Specifically, we will look at the induction of competence in Bacillus subtilis and the development of persister cells in E. coli. In both of these cases, the cells are genetically and environmentally identical. But in each case, a subset of the population undergoes a drastically different developmental change.
al environments can undergo very different developmental changes. Specifically, we will look at the induction of competence in Bacillus subtilis and the development of persister cells in E. coli. In both of these cases, the cells are genetically and environmentally identical. But in each case, a subset of the population undergoes a drastically different developmental change.As Bacillus begins to enter stationary phase, a subpopulation of cells begins to become competent, allowing a small percentage of cells to take up DNA from the environment. This process is activated by the ComK protein, which also activates its own expression in a positive feedback loop. As ComK levels increase, a threshold is hit, leading to a rapid increase in ComK production, and a subpopulation of cells begin to enter competence. This leads to the question as to the mechanism which allows only a subset of cells to enter competence.
Back in 2007, the Dubnau Lab at UMDNJ published a paper in Science discussing how very slight variations, or noise, in the amount of comK mRNA could lead to certain cells becoming competent, while others remained vegetative. The authors used fluorescent in situ hybridization to measure the exact amount of comK mRNA in individual cells. They found that in early stationary phase the number of comK mRNAs in the total population increased from 0.7 to 1 per cell. As the population average increased ever so slightly, a subpopulation began switching to competence. Thus, as the average was shifted, a small subset was well-enough above the mean to hit the ComK threshold and enter competence. Noise in gene expression dictated which cells became competent.
Those cell to cell variations of comK expression could be due to intrinsic noise, events such as mRNA decay or transcription initiation rates, or extrinsic noise such as the concentration of polymerases or transcription factors. To test which of these contributed to comK variability, the authors again used the in situ hybridization technique to measure comK mRNA as well as a control mRNA under a comK promoter. Extrinsic variations should affect both mRNAs equally, while intrinsic variations would only be acting on a single locus. Counting the number of mRNAs immediately before the induction of competence, they found that the number of each mRNA type was uncorrelated, indicating that random intrinsic noise was responsible for the variations in comK expression.
So what does this all mean?
In essence, we must first understand that when we see a population with our favorite gene (O
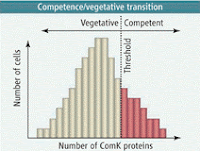 FG) expressed at level "X," really "X" just represents the mean expression of the population, and that per cell, the expression of this gene falls somewhere in a normal curve around "X." Cells with slightly higher or lower levels of OFG can end up having large effects on phenotypic outcomes, particularly if OFG is a regulator.
FG) expressed at level "X," really "X" just represents the mean expression of the population, and that per cell, the expression of this gene falls somewhere in a normal curve around "X." Cells with slightly higher or lower levels of OFG can end up having large effects on phenotypic outcomes, particularly if OFG is a regulator.Importantly, it shows that not all phenotypic outcomes are entirely the sum of genetics and environment, but can be due to completely random events within the cell.
It also raises the question as to why the whole population would only want a subset to become competent. Perhaps it is a form of the bacteria hedging their bets, allowing only a small percentage to take a great risk (i.e. bringing in harmful DNA, delaying sporulation) to receive great reward (new genetic information to outcompete others), while the remaining population continues laissez faire. But this can be a topic for next week when we take a look at bet-hedging in persister cells.
Source:
Maamar H, Raj A, & Dubnau D (2007). Noise in gene expression determines cell fate in Bacillus subtilis. Science (New York, N.Y.), 317 (5837), 526-9 PMID: 17569828
Other Articles of Interest:



2 comments:
I'm gonna have to disagree with your definition of environment. If you look at it from a physics point of view the environment is intrinsically stochastic. I don't see why you would consider noise/stochastic events not a part of the environment. While I do agree with you in principle that perhaps people have not recognized the importance of random events affecting phenotypes, I do not think that the word environment doesn't already cover those random events.
Ahh, this brings me back...
Justin
Post a Comment